-Search query
-Search result
Showing all 17 items for (author: gajhede & m)

EMDB-15985: 
Small molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: Development of screening assays and insight into GluK3 structure
Method: single particle / : Gajhede M, Boesen T

EMDB-15986: 
Ionotropic glutamate receptor K3 with glutamate and BPAM
Method: single particle / : Gajhede M, Boesen T

EMDB-15217: 
PAPP-A dimer in complex with a dimer of the inhibitor STC2
Method: single particle / : Kobbero SD, Oxvig C, Gajhede M, Boesen T

EMDB-15219: 
PAPP-A dimer in complex with endogenous STC2 inhibitor.
Method: single particle / : Kobbero SD, Oxvig C, Gajhede M, Boesen T

EMDB-15220: 
Partial dimer complex of PAPP-A and its inhibitor STC2
Method: single particle / : Kobbero SD, Gajhede M, Mirza OA, Boesen T, Oxvig C

EMDB-15221: 
PAPP-A dimer in complex with its inhibitor STC2
Method: single particle / : Kobbero SD, Gajhede M, Mirza OA, Boesen T, Oxvig C

PDB-8a7d: 
Partial dimer complex of PAPP-A and its inhibitor STC2
Method: single particle / : Kobbero SD, Gajhede M, Mirza OA, Boesen T, Oxvig C

PDB-8a7e: 
PAPP-A dimer in complex with its inhibitor STC2
Method: single particle / : Kobbero SD, Gajhede M, Mirza OA, Boesen T, Oxvig C

EMDB-4921: 
Density map of GluA2cryst with agonist AMPA, Class 1, compact
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4875: 
Density map of GluA2cryst in the apo state
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4920: 
Density map of GluA2cryst with negative allosteric modulator GYKI53655 and competitive antagonist ZK200775
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4922: 
Density map of GluA2cryst with allosteric modulator GYKI53655 and agonist AMPA, Class 2, loose
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4923: 
Density map of GluA2cryst with negative allosteric modulator GYKI53655
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4924: 
Density map of GluA2cryst with agonist AMPA, Class 2, loose
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS
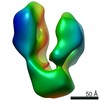
EMDB-4925: 
Density map of GluA2cryst with allosteric modulator GYKI53655 and agonist AMPA, Class 1, compact
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
